Mass Spectrometry & Spectroscopy
New Colour Measurement and Automation with UV/VIS Excellence Spectrophotometers
Nov 01 2017
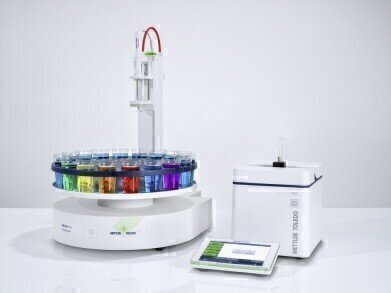
METTLER TOLEDO introduces the updated UV/VIS Excellence spectrophotometers with new features; colour measurement and the support of InMotion sample changers for custom automation.
Colour Measurement
The new UV/VIS Excellence instruments are able to measure the colour of samples in addition to the established optical performance. Spectra and colour can be measured simultaneously, streamlining the analysis. The most common color scales are supported, namely CIE L*a*b* and Luv, Tristimulus, Lab according to Hunter, Chromaticity (x, y), APHA, Pt/Co, Hazen, Yellowness Index, Saybolt, Gardner, EBC, ASBC and Hess-Ives. Mettler-Toledo's OneClick™ user interface completes the analysis of colour and other qualities of a sample within seconds. With the addition of LabX software, the toolbox is complete to create a compliant and data integrity lab solution.
Automation
Mettler-Toledo InMotion sample changers are used successfully in conjunction with several analytical instruments. With the new version of Excellence spectrophotometers, the InMotion sample changers can now also automate the analysis of large numbers of spectrophotometric samples. The samples are transferred to the instrument with a peristaltic pump, and measured in a flow-through cell; providing the user with a fully automated work-flow, including the cleaning steps.
LabX 2018
The new version of Excellence spectrophotometers is accompanied by the release of LabX 2018, which adds work-flow flexibility and supports compliance with regulations such as 21 CFR Pt 11. The combination of LabX 2018 and the updated spectrophotometers allow for complete customization of UV/VIS workstations to the workflow in the lab. The laboratory solution performs calculations, reports, and data-management, saving time for operators and lab managers, and reducing the risk of transcription errors.
For more information please click here.
Digital Edition
Lab Asia 31.2 April 2024
April 2024
In This Edition Chromatography Articles - Approaches to troubleshooting an SPE method for the analysis of oligonucleotides (pt i) - High-precision liquid flow processes demand full fluidic c...
View all digital editions
Events
May 21 2024 Lagos, Nigeria
May 22 2024 Basel, Switzerland
Scientific Laboratory Show & Conference 2024
May 22 2024 Nottingham, UK
May 23 2024 Beijing, China
May 28 2024 Tel Aviv, Israel




.jpg)